Infer Metabolic Enzyme Activity From Gene Expression Profile
An online platform for predicting metabolic enzyme activity, stratifying metabolic subtypes of cancers, and exploring their clinical relevance.
Background: Metabolic enzyme activity plays a key role in controlling the conversion rates of metabolic reactions. However, accurately and efficiently measuring enzyme activity on a large scale remains a challenge due to the complex, multi-level regulation involved. To overcome this, we developed a computational workflow called iMetAct (https://github.com/xiaolab-xjtu/iMetAct), which infers metabolic enzyme and pathway activity using widely accessible gene expression data. Detailed explanations of the core principles behind iMetAct, along with its performance evaluation, are provided in our manuscript.
MoreCancer Stratification
Exploring metabolic subtypes of 33 TCGA cancer types.
Clinical Characteristics
Analysis of individual metabolic enzymes.
Subtype Prediction
ML-based forecasting of HCC metabolic subtypes.
Activity Inference
Upload user expression profile and evaluate samples' enzyme activity.
Data Download
Metabolic enzyme activity for each TCGA cancer type.
What can our web do!
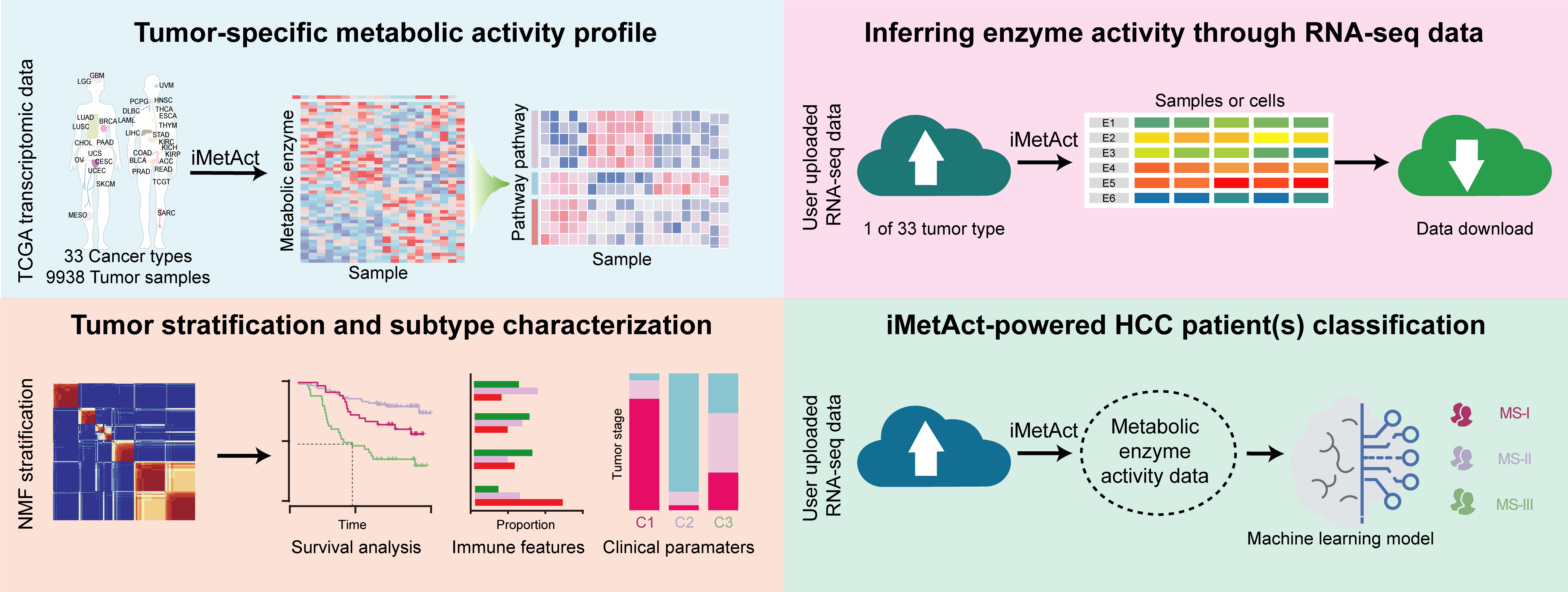
Tumor Metabolic Enzyme Activity Clustering and Results Display
This section of the website enables users to select tumors of interest. Through our precomputed tumor metabolic enzyme activity data and data clustered using the Non-negative Matrix Factorization (NMF) method, combined with NMF algorithms, users can explore the metabolic characteristics of various subtypes of the selected tumor. Additionally, this includes differences in survival rates and immune features among the subtypes. Users have the capability to download completed figures for their analysis and presentations.Step1: Select the tumor of interest and choose the algorithm used for NMF clustering.
Based on multiple index results from the NMF method, determine the optimal number of clusters.
Step2:This feature allows you to select the number of clusters needed for your analysis.
Reactome Metabolic Pathways
CIBERSORT Immune Cell Proportion
Relationship Between Metabolic Enzymes and Patient Prognosis
The Survival Analysis section of this webpage allows users to select a tumor type and specific metabolic enzymes of interest. We then generate survival curves based on the clustering of metabolic enzyme activity within the chosen tumor. The Survival Map feature enables users to compare survival differences for a selected metabolic enzyme across 33 different types of tumors.Hepatocellular Carcinoma Metabolic Subtype Prediction
This section of the website allows users to upload their own Hepatocellular Carcinoma (HCC) RNA-seq data. We will compute the metabolic enzyme activity and predict the metabolic subtypes of the samples. The metabolic and clinical characteristics of each subtype can be referenced in our publication.
Your sample(s) predicted result:
Evulate Tumor Metabolic Enzyme Activity
This section of the website allows users to upload their own tumor datasets. We will compute the metabolic enzyme activity of the user's samples using the iMetAct algorithm. However, it is important to note that users must select the same type of tumor as their data to obtain accurate metabolic enzyme activity results.Evaluated result
The generated data in this study can be downloaded.
Supplementary materials along with our study.
Supplementary tables
Download single TCGA cancer metabolic enzyme activity.
Download all TCGA cancer metabolic enzyme activity based on our calculated.
